Comprehensive mouse microbiota genome catalog reveals major difference to its human counterpart
Abstract
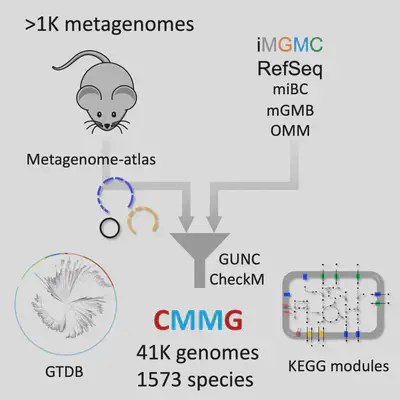
Mouse is the most used model for studying the impact of microbiota on its host, but the rep- ertoire of species from the mouse gut microbiome remains largely unknown. Accordingly, the similarity between human and mouse microbiomes at a low taxonomic level is not clear. We construct a comprehensive mouse microbiota genome (CMMG) catalog by assembling all currently available mouse gut metagenomes and combining them with published refer- ence and metagenome-assembled genomes. The 41’798 genomes cluster into 1’573 spe- cies, of which 78.1% are uncultured, and we discovered 226 new genera, seven new families, and one new order. CMMG enables an unprecedented coverage of the mouse gut microbiome exceeding 86%, increases the mapping rate over four-fold, and allows func- tional microbiota analyses of human and mouse linking them to the driver species. Compar- ing CMMG to microbiota from the unified human gastrointestinal genomes shows an overlap of 62% at the genus but only 10% at the species level, demonstrating that human and mouse gut microbiota are largely distinct. CMMG contains the most comprehensive col- lection of consistently functionally annotated species of the mouse and human microbiome to date, setting the ground for analysis of new and reanalysis of existing datasets at an unprecedented depth.
Awards
The publication was selected as Remarkable output 2022 by the Swiss bioinformatic institute.
What the committee said about the work:
“An excellent functional and taxonomic summary of the mouse gut metagenome, built using a solid bioinformatics pipeline. This is a great resource for researchers translating results from mouse to human.”
“This is a great resource for researchers translating results from mouse to human.”
— SIB (@ISBSIB) April 13, 2023
🥁 That’s what the committee said about this work, one of the #SIBRemarkableOutputs 2022 👏
👉 Find out more: https://t.co/pJp1rtnDDc#microbiome #genomics pic.twitter.com/DSHGbaj0y9