Binning with more than one Sample
Recent years we saw an explosion of genomes beeing assembled from megagenomes. Most of the studies assemble and bin each sample individually, because this is the most scalable. Maybe it’s time to think if this approach is really the best?
In a recent article Jennifer Mattock and Mick Watson1 makes a definitive case about the advantages of using multiple samples for metagenomic binning. MAGs assembled from single samples contain often hidden contamination. Contamination that is not detected by checkM & co, but is clerly vissible ussing a cross-sample correlation. Using the Co-abundance information one can discriminate even between closely related species that would be merged together by single-sample binning. One one hand this is not new. We know that co-abundance is usefull for binning. But on the other hand they clearly drive the point home.
Usually, in bioinformatics methods development, we rarely see that one method is clearly outperforms all other methods. Often benchmarks present a list of good tools and the user has to decide which method is the best for his/her data. Therefore I was quite surprises to read the following statement at the end of the abstarct:
While resource expensive, multi-coverage binning is a superior approach and should always be performed over single-coverage binning.
Ok, now let’s use multiple sample for binning. Inspired by this article I set co-abundance binning as the default in Metagenome-Atlas v2.18.
It is possible to map the reads from each sample to the contigs of all other samples and use this for binning. However, this this cross-mapping is quite ressource intensive.

Some would suggest this is the best approach and don’t care about scalability, but there is another approach: co-binning, also called multi-split by the authors who first used it2 .

For co-binning we concatenate contigs from multiple samples and map all the reads to these combined contigs. This way we improve computational efficiency, but still get the benefits of multi-sample binning. This approach worked very good in the variational autoencoders for metagenomic binning (VAMB)2. If it holds up to rigorous assessment by Mattock and Watson remains to be seen.
Also there are many other questions still open. For example, should we count multi-mapping for the co-binning approach or to what level of identity should it be filtered? Using unfiltered mapping will likely map reads from different species. Don’t taking into account multi-mapping will likely lead to random distribution of the reads and so to weakinging of the co-abundance signal.
What is next?
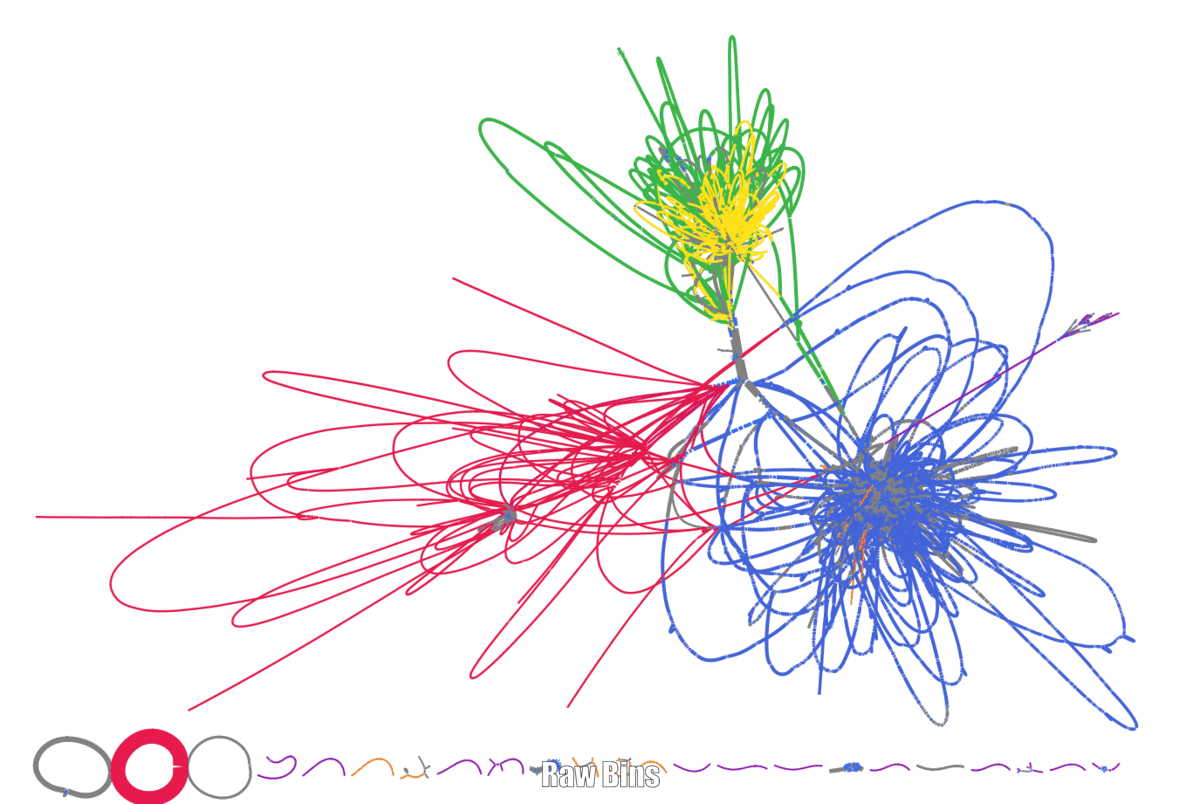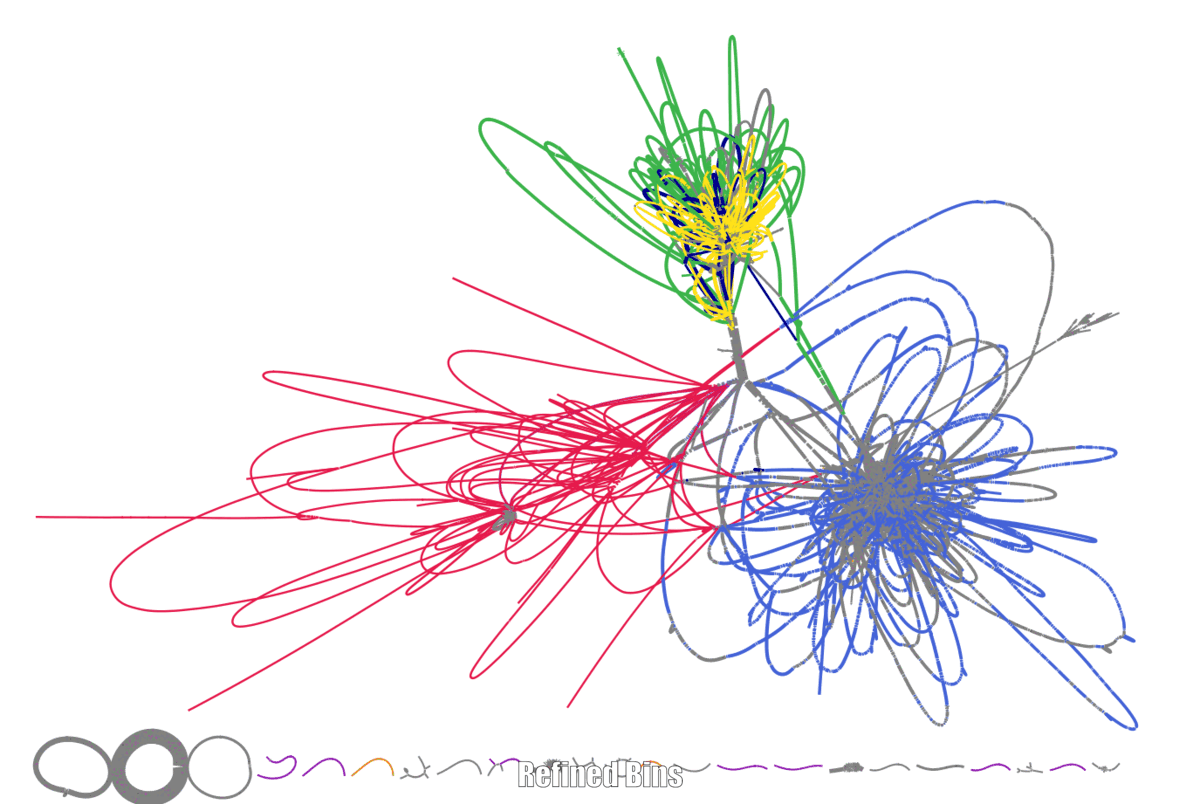
If we are already here to think about what could improve metagenomic binning. Then Using the Assembly graph(s) is a logical next step. Different algorithms have been proposed to use the assembly graph for binning.
Here are my first experiments with using binspreader3 for a small metagenome dataset.
References
1. Mattock, J. & Watson, M. A comparison of single-coverage and multi-coverage metagenomic binning reveals extensive hidden contamination. Nature Methods 20, 1170–1173 (2023).
2. Nissen, J. N. et al. Improved metagenome binning and assembly using deep variational autoencoders. Nature Biotechnology 39, 555–560 (2021).
3. Tolstoganov, I., Kamenev, Y., Kruglikov, R., Ochkalova, S. & Korobeynikov, A. BinSPreader: Refine binning results for fuller MAG reconstruction. iScience 25, 104770 (2022).